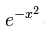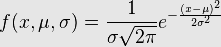Central Limit Theorem and the Gaussian/Normal distribution
The Gaussian distribution is an extremely popular distribution for most things in statistics related to population studies and biology in general. However, for the longest amount of time I did not really understand why it was really important. The Central Limit Theorem was a buzzword that I didn't have a feel for. However, using R it became very intuitive.
The CLT states that the mean/average/sum or additive effects of a large number of independent random events occurring with uniform probability will converge towards a bell shaped curve called the Gaussian or Normal distribution.
To see this in R:
unifmat<-matrix(runif(1000000),1000,1000)
hist(as.vector(unifmat))
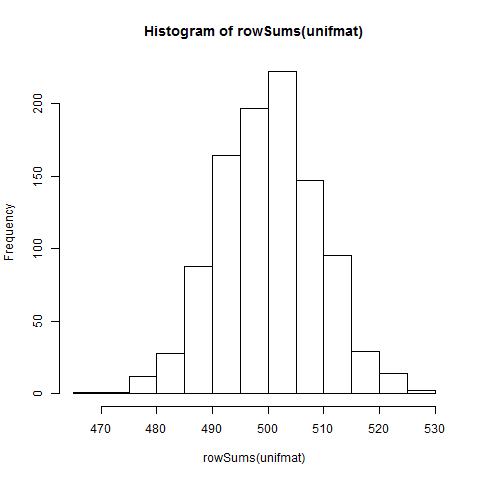
Now that is a uniformly distributed set of numbers that we have there inside that matrix that we collapsed to a vector and saw with a histogram that it was pretty much uniformly distributed. Now we take the averages/sums and see what the distribution is like
hist(rowSums(unifmat))
hist(rowMeans(unifmat))
Both of them look the same because the mean of the rows is just a scaled-down-by-the-same-number version of the sums of the rows.
So the Central Limit Theorem works in simulation. In practice, the distribution of heights in a population, which is believed to be a multi-loci inherited complex trait, is normally distributed presumably because the recombination of genes is a random event. That means most people are of a certain height and the shorter and the taller ones become rarer as you go to the left or the right of the X-axis. It is possible to estimate the proportion/percentage of people who will be taller than average by estimating the area under the curve for a certain height range divided by the total area under the curve. This distribution holds for IQ's as well. The reason why the bell curve distribution holds is because the mixing is uniformly random. If you start selecting from specific areas of the curve and then generating means then you will get a skew towards the left or the right. Now think Francis Galton, regression to the mean and Eugenics.
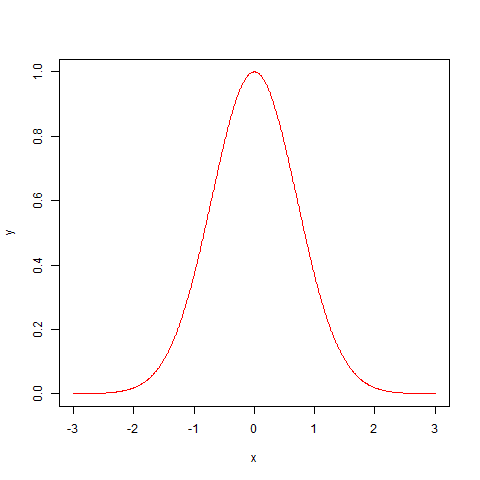
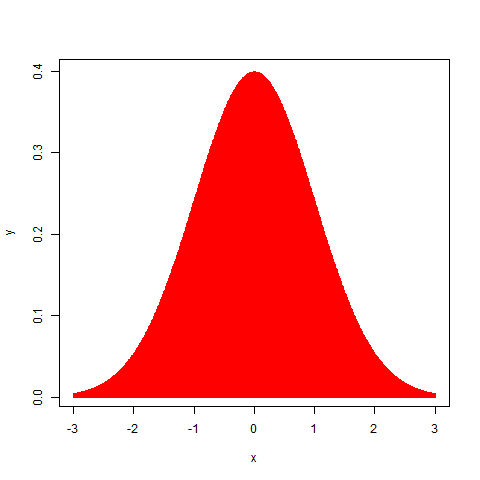
Now, we will generate this distribution using an analytic formula that allows us to feed in numbers and get a nice smooth Gaussian curve.
x=seq(-3,3,by=0.01)
y=exp(1)^(-1*x^2)
plot(x,y,type="l",col="red")
This comes from
Now the normalized version of this curve has an area of 1 under it. The reason for insisting on an area of 1 is that statisticians, especially the ones who love probability like to describe phenomena in terms of what they called probability distribution functions (PDF). Now these probability loving statisticians would die if the sum of all the probabilities would not add up to 1. So the area under curve (AUC) is one and it has been done by transforming the formula into something that looks like this.
We can code our own R function to generate Gaussian curves and this is how we will go about it.
Gauss<-function(x,mu,sigma){
gx<-(1/(sigma*sqrt(2*pi))) * exp(1)^(-1*((x-mu)^2/(2*sigma)))
return(gx)
}
Now to use it:
x<-seq(-3,3,by=0.01)
y<-Gauss(x,0,1) # we pass x and the mu/mean and the sigma/standard deviation
plot(x,y,type="h",col="red")
So, with that we have our own Gaussian function to play with. Especially now, we can modify the mean and the variance and obtain our own custom shaped normal distributions. We can also take two curves and perform math operations like adding, multiplying and other things with them to see what happens to their shapes.
First thing to do is to make a Dirac delta function, an extremely thin normal distribution that looks like a spike.
y=Gauss(x,0,0.001)
plot(x,y, pch="*",col="blue",type="b")
So you can create your own thin/fat curves by tweaking the sigma/variance, to shift the curve to the left or right of zero, simply choose a non-zero, negative or positive mean. Since this post is approaching the TL;DR Limit, I will put the experiments with Gaussians in another post.